Translate this page into:
Poly(A)-Specific Ribonuclease Deficiency Leads to Deregulated Expression of Genes Involved in Sex Determination and Gonadal Maturation in Zebrafish
Address for correspondence Anirban Chakraborty, PhD, Division of Molecular Genetics and Cancer, Nitte University Centre for Science Education and Research (NUCSER), Paneer Campus, Kotekar-Beeri Road, Nitte (Deemed to be University), Deralakatte, Mangaluru, Karnataka-575018, India (e-mail: anirban@nitte.edu.ina).
This article was originally published by Thieme Medical and Scientific Publishers Pvt. Ltd. and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Background
Deadenylation, the process of removal of poly (A) tail of messenger ribonucleic acids (mRNAs), is a rate-limiting step in mRNA stability, and poly(A)-specific ribonuclease (PARN) is the most important exonuclease involved in this process. Besides mRNA stability, PARN is also involved in several other processes including telomere maintenance, noncoding RNA maturation, ribosome biogenesis, and TP53 function. Previously, we have shown that zebrafish PARN null mutants are viable and fertile but turn out to only develop into males, indicating a role in oogenesis. The present study was focused on analyzing the expression of genes involved in sex determination and gonadal development in PARN mutant zebrafish.
Materials and Methods
Total RNA was extracted and quantitative real-time polymerase chain reaction was performed to determine the expression level of genes involved in gonad development in PARN mutant embryos (4 days postfertilization [dpf]) and adults (120 dpf) in comparison to their wild-type siblings. The expression levels were estimated by the AACT relative quantification method.
Results
At 4 dpf, the expression of germ cell-specific genes did not show any significant difference in the null mutants compared to the heterozygous and their wild-type siblings, suggesting no effect on germ cell differentiation due to the loss of PARN. However, the majority of the ovary-associated genes analyzed showed an increased expression in PARN null and heterozygous mutants compared to the wild-type siblings. Intriguingly, the expression of testis-associated genes showed decreased expression in the mutants compared to their wild-type siblings at 4 dpf. In adult stages, as expected, the expression of genes that jointly regulate the proper formation and function of ovaries and testes showed decreased expression in PARN null mutants. Interestingly, the expression of genes involved in the differentiation of testes, despite showing a decreased expression in the mutants, was comparable between the null and heterozygous mutants.
Conclusion
Taken together, these results suggest that the loss of PARN does not affect germ cell differentiation but affects the sexual differentiation that happens at later stages of development, particularly the process of oogenesis, in zebrafish.
Keywords
exonuclease
mRNA stability
PARN
gonadal genes
oogenesis
spermatogenesis
Introduction
The turnover of messenger ribonucleic acid (mRNA) in a cell is a balance between its synthesis and its degradation, The process mRNA degradation in eukaryotic cells requires a systematic removal of the poly A tail called deadenylation.1 The poly(A)-specific RiboNuclease (PARN), a 74-kDa exonuclease belonging to the DEDD superfamily and first identified from HeLa cells in 1991, is considered the most critical enzyme in the process of deadenylation.2–4
In recent years, it has become evident that PARN has multiple other roles in the cell, not just mRNA stock clearing.5 PARN is involved in the maturation of H/ACA box snoRNAs,6 a crucial member of the telomerase RNA (TERC) complex, and these snoRNAs pseudouridylate rRNA as a part of the process of rRNA processing and maturation. PARN shows nucleolar localization,7 coprecipitates with pre-40S particles, and participates in the processing of the 3'-end of the human 18S rRNA.8 These observations point strongly to its role in ribosome biogenesis. Further, the discovery of mutations in PARN in diseases of telomere biology, particularly those that affect hematopoiesis, has led to our understanding that defects in 3′end trimming of mRNA can impair hematopoiesis, as shown in mice9 and zebrafish.10
Destabilization of maternal mRNAs is crucial for the activation of zygotic transcription. Indeed, a study on Xenopus has revealed that PARN is responsible for maternal mRNA degradation during oocyte maturation and embryogenesis.11
The process of sex determination and gonad development in zebrafish is complex and it takes about 90 days postfertilization (dpf) for complete maturation of ovary and testis. Zebrafish are born females and the first signs of an ovary-like structure are seen as early as 10 dpf. The process of sex differentiation occurs between 17 and 25 dpf where the gonads remain bipotential. While half of the embryos maintain juvenile oogenesis, a number of gene networks work in a coordinated manner to initiate the process of juvenile oocyte apoptosis in the remaining half, leading to the “ovary to the testis” gonadal transformation process and ultimately resulting in the formation of the testis. At 90 dpf, the ovary and the testis are sexually differentiated.12
Previously, we have shown that loss of PARN affects oogenesis in zebrafish and parn null mutant zebrafish, though develop normally as viable and fertile adults, turning out to be only males.13 To further understand the molecular basis of the tissue-specific phenotype that we observed in zebrafish, in the present study, we systematically examined the expression profiles of germ cell-specific genes (dnd1, dyrk1a, nanos1), ovary-associated genes (foxl2a, fanc1, nanos1, nanos2, cyp19ala, piwil1), testis-associated genes (sox9a, dmrt1, hsf5, ar), and other gonad-associated genes (brca2, cyp11c1, cyp17a1) in embryonic and adult stages of parn null mutants. The results suggest that the loss of PARN does not affect the germ cell differentiation at embryonic stage (4 dpf) but affects the sexual differentiation that happens at later stages (post 25 dpf) of development, particularly the process of oogenesis, which results in an all-male phenotype in null mutants.
Materials and Methods
Zebrafish Husbandry and Rearing
Adult zebrafish (AB line) used in this study were obtained from a multilinking recirculatory rearing facility with a temperature maintained at 28 ± 0.5°C and with a 14/10day/night cycle. The process of mating and collection of fertilized eggs were as per the standard protocols followed in zebrafish labs.13 The animal ethics approval was duly obtained before the commencement of the study (NGSMIPS/IAEC/Nov-2019/154).
Identification of PARN Mutants through Genotyping
The PARN mutants, generated through the CRISPR/Cas9 gene-editing tool,13 were used in this study. The zygosity of the fishes was determined through genotyping of deoxyribonucleic acid (DNA) obtained from tail clips. The process of tail clipping involved anesthetizing the fishes by 0.02% MS-222 (Tricaine), transferring immediately onto a petri dish using a plastic spoon, and clipping a part of the caudal fin with a sterile blade. The clipped tail was immediately put into lysis buffer containing 10 mM tris HCl, 50 mM KCl, 0.3% tween20, 0.3% NP40, and 1 mM ethylenediaminetetraacetic acid, and denatured at 98°C for 10 minutes. Denaturation was followed by the addition of 2 mg/mL proteinase K and incubation of the sample at 55°C overnight. The next morning, the reaction was stopped by incubating the reaction mixture at 98°C for 10 minutes. The crude DNA thus obtained was diluted 1:5 and used as a template for polymerase chain reaction (PCR) and the target region was amplified. The sequence information of the PARN genotyping primers is mentioned in the Supplementary Table S1. Confirmation of the zygosity status of the mutants was done by manual analysis of the Sanger sequence of the PCR product.
Analysis of Sanger Sequencing Data
To check for 5 base pair del heterozygous/homozygous PARN mutation, the software called Poly Peak Parser was used (http://yosttools.genetics.utah.edu/PolyPeakParser/). This software makes an alignment of the query to the wild-type sequence and, in the process, it identifies the gaps in the sequence.
Gene Expression Analysis by Real-Time PCR
RNeasy Minikit (Qiagen, Germany) was used for total RNA extraction from different tissues of adult parn mutants and their wild-type siblings. In the case of embryos, RNA was extracted from the whole animal (day 4). The conversion of RNA to complementary DNA (cDNA) was carried out using commercial kits (PrimeScript 1st strand cDNA Synthesis Kit; Takara, Japan). The expression level of genes involved in gonad development in the mutants and their wild-type siblings was carried out by quantitative real-time PCR (QuantStudio3, Thermo Fisher Scientific, United States). The expression levels were estimated by the ΔΔCT relative quantification method. The gene elfa was used as an internal control. The sequences of the primers are given in Supplementary Table S1.
Statistical Analysis
Microsoft Excel 2010 and GraphPad Prism 8.4.3 were used for all statistical analysis. The experiments were carried out in triplicates and a p-value of < 0.05 was considered statistically significant.
Results
PARN Null Mutant Zebrafish Shows Deregulated Expression of Genes Involved in Sex Determination and Gonadal Maturation
Since PARN deficiency impacted oogenesis in zebrafish,14 we assessed the expression of genes involved in sex determination and gonadal maturation using gonadal tissues of parn null mutant zebrafish by quantitative reverse transcriptase-PCR and compared with that in heterozygous zebrafish relative to their expression in the wild-type siblings. The expression of gonad-associated genes was checked at two developmental stages; embryonic stage (4 dpf) and adult stage (120 dpf). While gonad tissues (ovary and testis) were used in adults for total RNA extraction, the whole animal was used for total RNA extraction in embryonic stages.
In adults, both testis-associated (hsf5, dmrt1, sox9a, ar) and ovary-associated (foxl2, fanc1, piwil1) genes were analyzed. The results indicated that loss of PARN resulted in decreased expression of all the testis-associated genes in the null mutant (PARN−/−) and heterozygous (PARN+/−) zebrafish compared to wild-type zebrafish (PARN+/+) except for ar that showed no significant difference between the wild-type and the heterozygous mutant zebrafish (Fig. 1A).

- Relative expression of testis and ovary-associated genes in parn null (PARN−/−) and heterozygous (PARN+/−) adults compared to their wild-type (PARN+/+) siblings, normalized to internal control elfa1 gene. Each histogram represents the expression of testis (A) and ovary (B)-specific genes, calculated as fold change compared to the expression in wild-type. **** indicates p < 0.0001, *** indicates p < 0.005, * indicates p < 0.05, ns indicates nonsignificant.
However, in the case of ovary-associated genes, since the null mutant zebrafish did not develop ovaries, only PARN+/− was compared to that of wild-type embryos. Among the three genes tested, only 2foxl2a showed decreased mRNA expression at statistically significant levels, while the other two genes (fancl and piwil1) did not show any difference (Fig. 1B).
At the embryonic stage, two maternally expressed germ cell-specific genes (dyrk1a, nanos1) showed decreased expression in both null and heterozygous mutants compared to the wild-type siblings, whereas the dnd1 transcript level was almost the same as the wild-type (Fig. 2A). However, when compared between parn null and heterozygous mutants, of the three genes checked, two genes namely dnd1 and nanos1 did not show any difference in their expression (Fig. 2A). As shown in our earlier study,14 heterozygous mutants developed as normal males and females, suggesting that loss of PARN has no effect on germ cell differentiation. As for the ovary-associated genes (foxl2, fancl, nanos1, nanos2, cyp19a1a) analyzed, three genes (foxl2, fancl, and nanos2) showed an upregulation in both null and heterozygous mutant zebrafish (Fig. 2B). In case of the other two genes, nanos1 showed a decreased expression in both null and heterozygous mutants compared to the wild-type embryos. However, there was no difference in expression when compared between the PARN null and heterozygous mutant zebrafish (Fig. 2B). The expression of cyp19a1a did not show any difference between the wild-type, null, and heterozygous mutants (Fig. 2B). Interestingly, the expression of both the testis-associated genes analyzed (sox9a, dmrt), was decreased in PARN null and heterozygous mutants at a statistically significant level when compared to the wild-type embryos (Fig. 2C). In addition, three more genes, namely, brca2, cyp17a1, and cyp11c1 that play an important role in the development of both male and female zebrafish were also analyzed and the expression of all these three genes was significantly decreased in parn null and heterozygous mutants, compared to the wild-type siblings (Fig. 2D). Taken together, these results suggest that the loss of PARN might not affect germ cell differentiation but it does affect the sexual differentiation that happens at later stages (>25 dpf) of development.
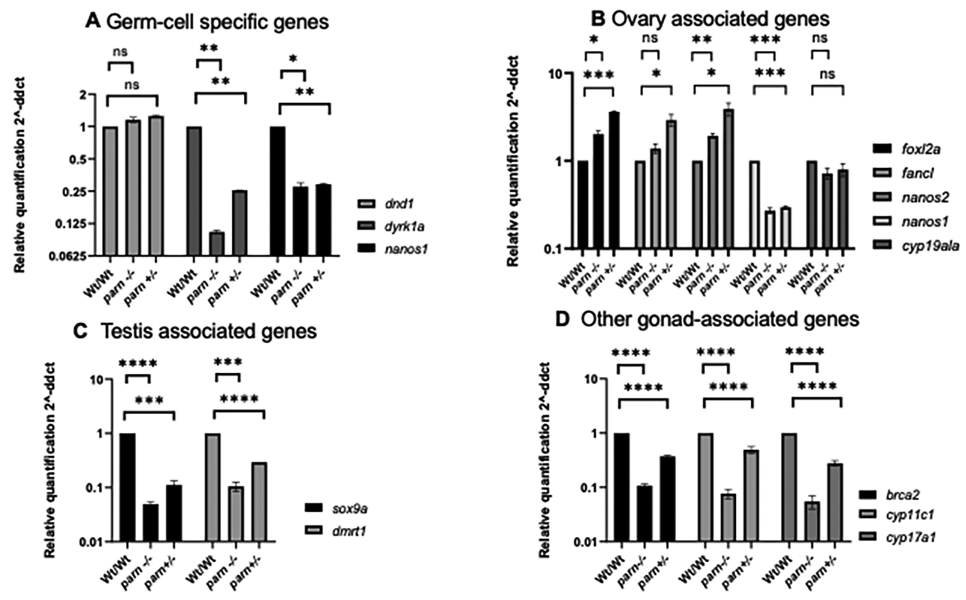
- Relative expression of germ cell-specific, testis and ovary-associated genes in parn null (parn−/−) and heterozygous (parn+/+) embryos compared to their wild-type (parn+/+) siblings, normalized to internal control elfal gene. Each histogram represents the expression of germ cell-associated genes (A), ovary-associated genes (B), and testis-associated genes (C), and other gonad-associated genes (D), calculated as fold change compared to the expression in wild-type. **** indicates p < 0.0001.
Discussion and Conclusion
The canonical function of PARN is its ability to remove adenosine residues in the 3′end of mature mRNA and hence it is crucial for efficient functioning of the mRNA decay pathway. However, several noncanonical functions of PARN have now emerged including a role in ribosome biogenesis, decay of noncoding RNAs,15 regulation of TP53,16,17 and maintenance of telomere complex.18,19 Intriguingly, mutations in PARN have been identified in several diseases that manifest as telomere dysfunction and bone marrow failure.20–22 Thus, in order to completely understand the conventional and emerging functions of PARN, loss-of-function studies, ideally in multicellular animal models are necessary to delineate the role of PARN in cellular processes.
Since the parn null mutants, described in our previous study,14 showed a gonadal phenotype, in this study we analyzed the transcript level of genes involved in sex determination and gonadal maturation at different stages of development14,23–25 in parn null mutants. At the embryonic stage, there was decreased expression of two of the three germ cell-specific genes analyzed, in both parn null and heterozygous mutants compared to their wild-type siblings, but without any significant difference between the two genotypes. In adult stages, as expected, the expression of genes that jointly regulate the proper formation and function of ovaries were downregulated in parn null mutants. Given the fact that the parn null mutants are fertile males,14 the decreased expression of genes involved in the differentiation of testes suggests no effect on the maturation of the testis.
As seen in PARN null mutants, atm and fancl null zebrafish also show an all-male phenotype ATP53-mediated apoptotic pathway in the primordial germ cells and has been suggested as the possible mechanism in these mutants. Interestingly, an earlier cell line-based study from our group has shown that the TP53 is downregulated in PARN-deficient conditions.26 Thus, it seems unlikely that TP53 is responsible for impaired oogenesis in zebrafish parn null mutants as seen in atm and fancl null mutants. Decay of maternal mRNAs that support early embryonic development is an important step during the activation of zygotic transcription. Indeed, PARN has been shown to be involved in inactivation of maternal mRNAs in Xenopus.27 Thus, it is conceivable that PARN deficiency leads to a defect in maternal mRNA decay, which in turn delays the process of zygotic transcription, resulting in deregulation in the process of gonad differentiation in PARN null mutant zebrafish. Whole transcriptome-based study at different time points during zebrafish development would be required to support this hypothesis. Nevertheless, the results of this study suggest that PARN is important during sex differentiation in zebrafish, and its deficiency affects the process of sex differentiation, results in an allmale phenotype in the null mutants. Disorders of sexual development (DSDs) are congenital conditions that manifest as atypical development of gonads and anatomical sex. It would be interesting to see if PARN is mutated in DSDs and whether it shows differential expression in these disorders.
Acknowledgments
We would also like to extend our sincere gratitude to Patrick Sips and Hanna De Saffel, Centre for Medical Genetics, Ghent University, Belgium, for their invaluable support.
Funding
This study was partially funded by the DST-SERB, Government of India.
Conflict of Interest
None declared.
References
- Mechanisms of deadenylation-dependent decay. Wiley Interdiscip Rev RNA. 2011;2(02):167-183.
- [Google Scholar]
- A 54-kDa fragment of the Poly(A)-specific ribonuclease is an oligomeric, processive, and cap-interacting Poly(A)-specific 3′ exonuclease. J Biol Chem. 2000;275(31):24222-24230.
- [Google Scholar]
- The mRNA cap structure stimulates rate of poly(A) removal and amplifies processivity of degradation. J Biol Chem. 2001;276(30):27923-27929.
- [Google Scholar]
- In vitro deadenylation of mammalian mRNA by a HeLa cell 3′ exonuclease. EMBO J. 1991;10(10):3067-3071.
- [Google Scholar]
- Poly (A)-specific ribonuclease (PARN): More than just “mRNA stock clearing”. Life Sci. 2021;285:119953.
- [Google Scholar]
- Maturation of mammalian H/ACA box snoRNAs: PAPD5-dependent adenylation and PARN-dependent trimming. RNA. 2012;18(05):958-972.
- [Google Scholar]
- Poly(A)-specific ribonuclease regulates the processing of small-subunit rRNAs in human cells. Nucleic Acids Res. 2017;45(06):3437-3447.
- [Google Scholar]
- Poly(A)-specific ribonuclease is a nuclear ribosome biogenesis factor involved in human 18S rRNA maturation. Nucleic Acids Res. 2017;45(11):6822-6836.
- [Google Scholar]
- Targeted disruption of Zfp36l2, encoding a CCCH tandem zinc finger RNA-binding protein, results in defective hematopoiesis. Blood. 2009;114(12):2401-2410.
- [Google Scholar]
- cpsf1 is required for definitive HSC survival in zebrafish. Blood. 2011;117(15):3996-4007.
- [Google Scholar]
- The mechanism and regulation of deadenylation: identification and characterization of Xenopus PARN. RNA. 2001;7(06):875-886.
- [Google Scholar]
- Zebrafish as an emerging model to study gonad development. Comput Struct Biotechnol J. 2020;18:2373-2380.
- [Google Scholar]
- The Zebrafish Book In: A Guide for the Laboratory Use of Zebrafish (Danio rerio) (4th ed.). Eugene: University of Oregon Press; 2000.
- [Google Scholar]
- Poly (A)-specific ribonuclease deficiency impacts oogenesis in zebrafish. Sci Rep. 2023;13(01):10026.
- [Google Scholar]
- PARN modulates Y RNA stability and its 3′-end formation. Mol Cell Biol. 2017;37(20):e00264-e17.
- [Google Scholar]
- Nuclear Tau, p53 and Pin1 regulate PARN-mediated deadenylation and gene expression. Front Mol Neurosci. 2019;12:242.
- [Google Scholar]
- The RNase PARN controls the levels of specific miRNAs that contribute to p53 regulation. Mol Cell. 2019;73(06):1204-1216.e4.
- [Google Scholar]
- Mutations in thes Poly (A)-Specific Ribonuclease (PARN) Impair Telomerase RNA 3′End Maturation in Dyskeratosis Congenita Patients. Washington, DC: American Society of Hematology; 2015.
- Poly(A)-specific ribonuclease deficiency impacts telomere biology and causes dyskeratosis congenita. J Clin Invest. 2015;125(05):2151-2160.
- [Google Scholar]
- Bone marrow failure and developmental delay caused by mutations in poly(A)-specific ribonuclease (PARN) J Med Genet. 2015;52(11):738-748.
- [Google Scholar]
- NCI DCEG Cancer Genomics Research Laboratory NCI DCEG Cancer Sequencing Working Group. Hoyeraal-Hreidarsson syndrome due to PARN mutations: fourteen years of follow-up. Pediatr Neurol. 2016;56:62-68.e1.
- [Google Scholar]
- Multiple bilateral hip fractures in a patient with dyskeratosis congenita caused by a novel mutation in the PARN gene. Osteoporos Int. 2021;32(06):1227-1231.
- [Google Scholar]
- Regulation of zebrafish gonadal sex differentiation. AIMS Mol Sci. 2016;3(04):567-584.
- [Google Scholar]
- PARN knockdown in cell lines results in differential and cell-specific alterations in the expression of cancer-associated mRNAs. Asian Pac J Cancer Prev. 2022;23(01):207-215.
- [Google Scholar]
- Nonsense mutations in close proximity to the initiation codon fail to trigger full nonsense-mediated mRNA decay. J Biol Chem. 2004;279(31):32170-32180.
- [Google Scholar]







